
How to use Plant Genome Editing Database
1. About Plant Genome Editing Database
This database provides information about plants that have been generated using CRISPR/Cas9 technology in order to study interesting and in many cases economically important traits. Information provided includes details of the transformation experiment, the name of the transformed plant variety, the DNA construct used including the guide RNA sequence and primers used to characterize resulting mutations, and details about the mutant plant line including the altered gene sequence, whether it is heterozygous or homozygous, and any phenotypes that have been observed (Fig. 1). Users are encouraged to make information available about their own CRISPR-generated plant lines and instructions are provided about how data can be submitted for inclusion in the database.
The database will be updated frequently. New dataset submission or publications related to genes mutated by CRISPR/Cas in the database will be released in "News" on the homepage.
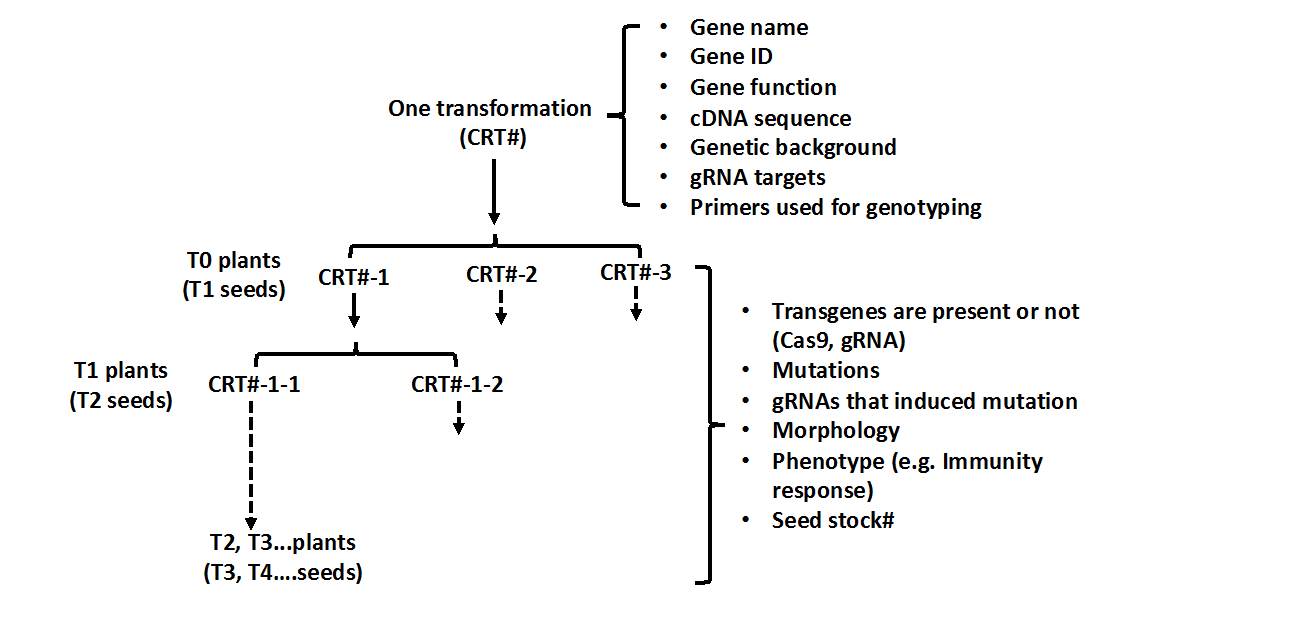 Fig. 1 Information provided for each transformant and their later generations in Plant Genome Editing database.
Fig. 1 Information provided for each transformant and their later generations in Plant Genome Editing database.
2. Search for specific CRISPR-generated mutations
Users begin by searching the database in three ways for genes that carry mutations (Fig. 2). a) "Search Gene" by using specific gene identifiers (e.g., Solyc10g085990) or the gene description; b) "Batch Query" by inputting a list of gene IDs; or c) "Search gRNA" by using specific gRNA sequences.
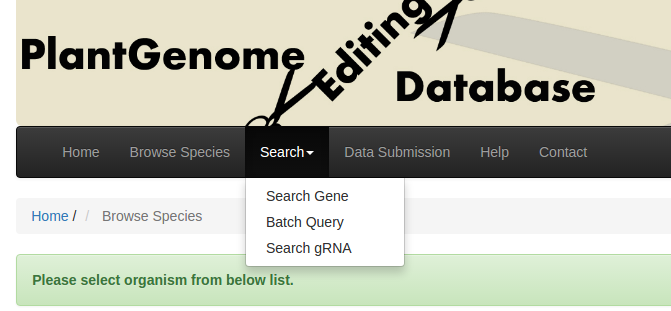 Fig. 2 Search for genes of interest.
Fig. 2 Search for genes of interest.
Or, users can begin by first choosing the plant species they are interested in and browsing the genes that carry mutations only in that plant species. Upon choosing the species, users will be guided to "List of Genes", which displays all the genes edited by CRISPR/Cas in this species (Fig. 3).
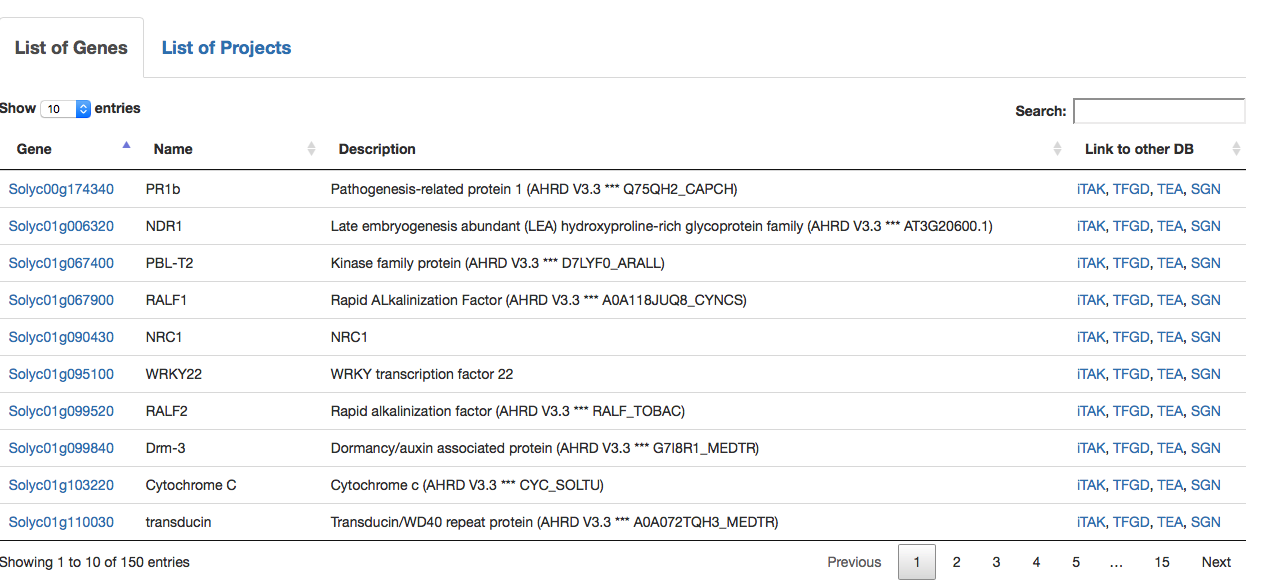 Fig. 3 List of genes in a selected plant species (e.g., in this case tomato).
Fig. 3 List of genes in a selected plant species (e.g., in this case tomato).
If a user clicks "List of Projects" (Fig. 3), details of the projects from which CRISPR data are provided is described (Fig. 4). By choosing a specific project, users can browse the list of genes and plant mutants only for the selected project.
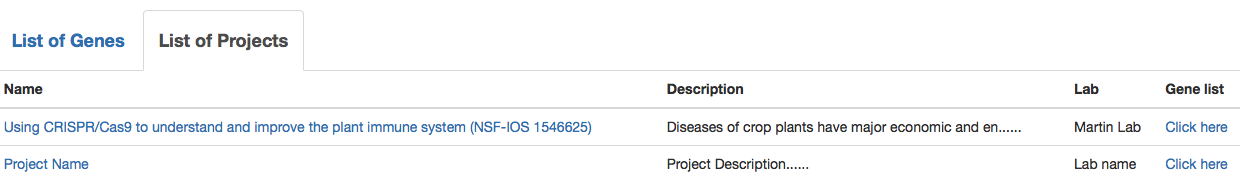 Fig. 4 List of projects in a selected plant species.
Fig. 4 List of projects in a selected plant species.
3. Information provided for each plant mutant
Upon selecting a gene of interest, users will see all information provided including gene name, gene function, the sequence & structure, a list of plants carrying mutations of this gene, and details of each plant mutant (genetic background, gRNA, mutation, phenotype, etc.) (Fig. 5).
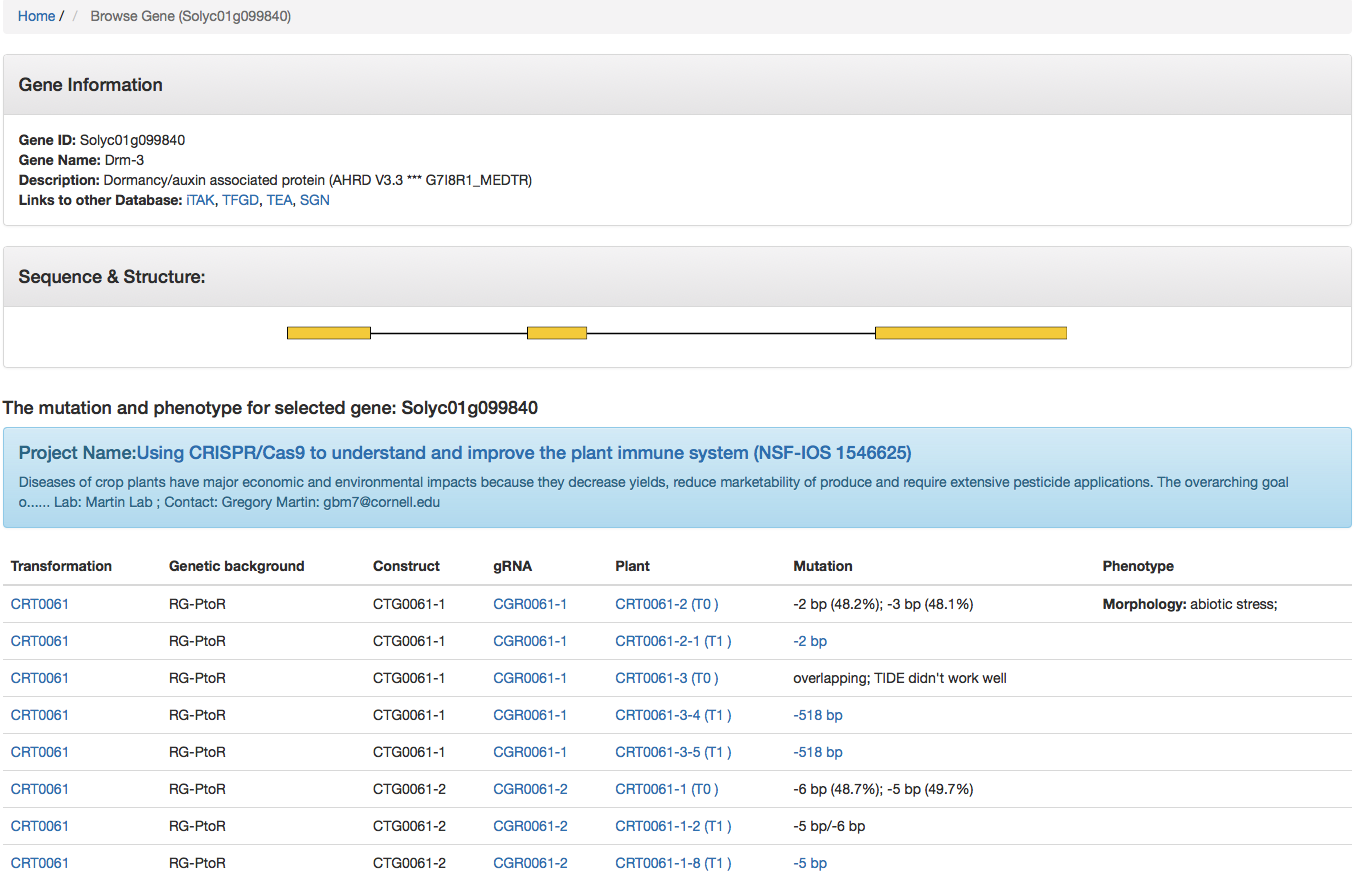 Fig. 5 Information provided for CRISPR mutants for gene Solyc01g099840.
Fig. 5 Information provided for CRISPR mutants for gene Solyc01g099840.
Users can click on a specific "Transformation #", "gRNA #" or "Plant #" to get more information (Fig. 5). For example, by clicking the transformation CRT0061, constructs used for this transformation (Fig. 6A) and a family tree with plants carrying targeted mutations are displayed (Fig. 6C). By clicking the gRNA CGR0061-1, the 20-nt gRNA sequence, primers used for genotyping the targeted mutation, and the project and transformation that the gRNA were used for are listed.
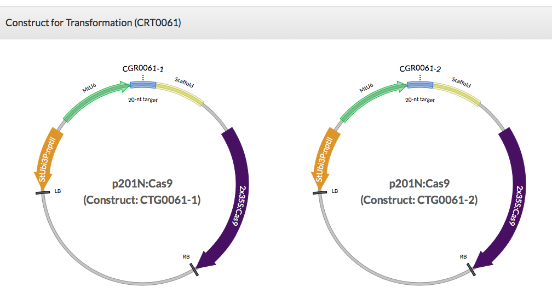
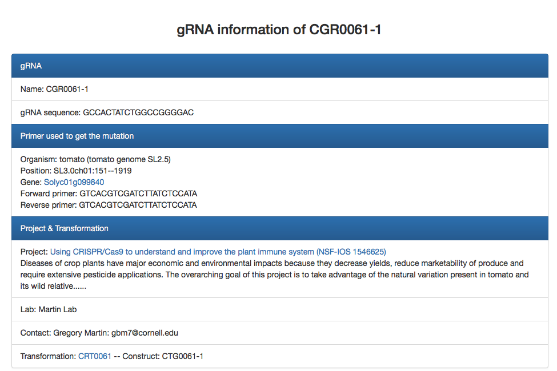
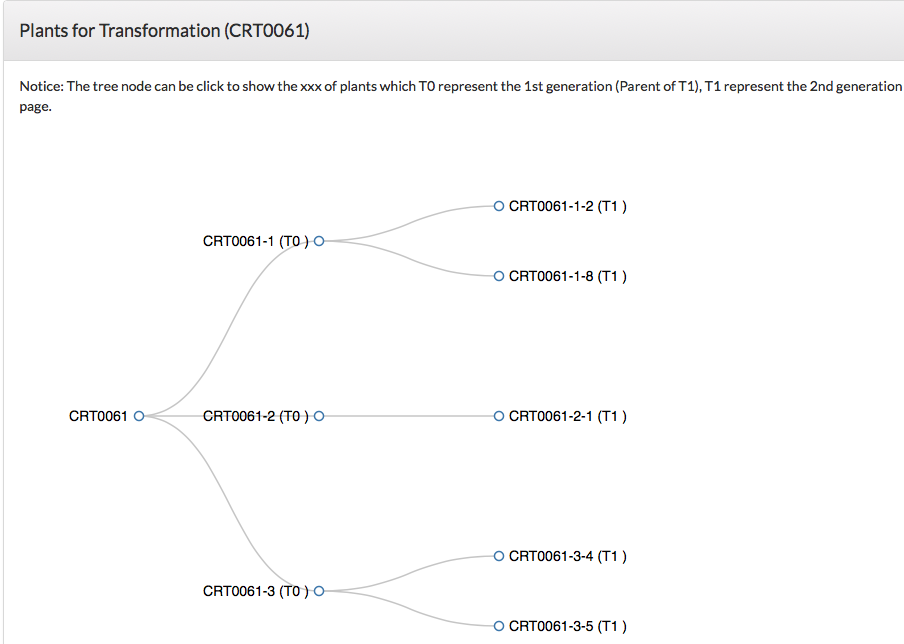
If a user clicks on a specific plant mutant (CRT0001-9-1), more details including the modified genes in this plant, gRNA target and mutation of each gene, and seed stock# are provided. If seeds are available, the user can contact the depositor to request the seeds for their own experimental (non-commercial) use (Fig. 7).
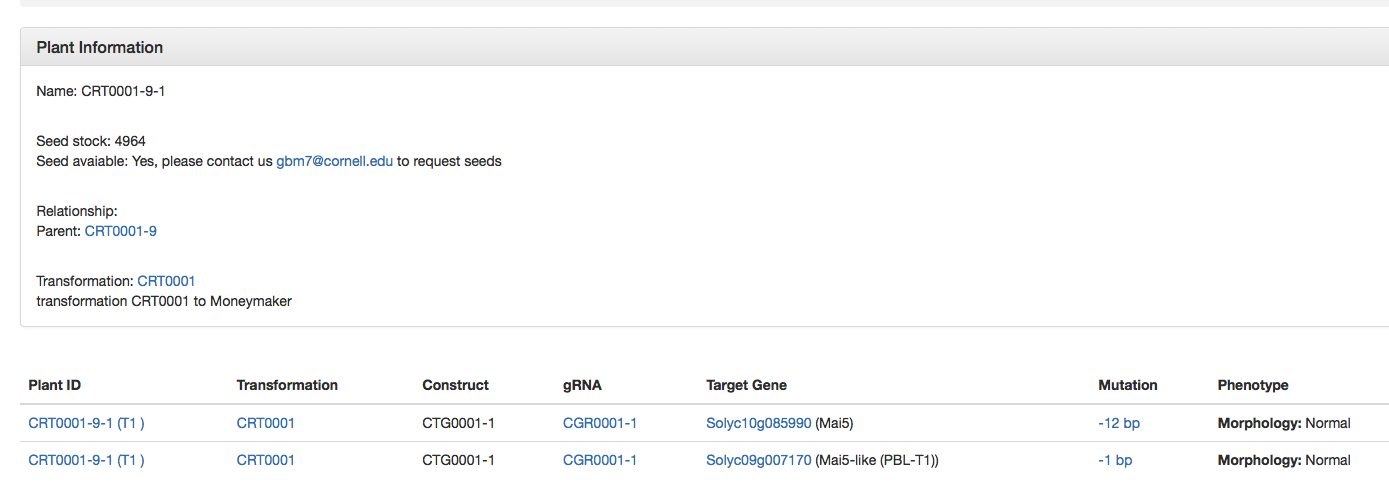 Fig. 7 Information displayed for each single plant carrying targeted mutations.
Fig. 7 Information displayed for each single plant carrying targeted mutations.For plants with homozygous (but not biallelic) mutations, clicking the mutation (such as -1 bp in Fig. 8) will lead to a page showing the mutation type in the genome region of the gene (Fig. 8; mutation is highlighted in purple, gRNA is highlighted in blue and exon is highlighted in yellow).
 Fig. 8 An example of mutation alignment.
Fig. 8 An example of mutation alignment.4. Submit your own CRISPR mutant data
Please check the Data Submission page on how to submit your own data. The submission page provides a step-by-step guide for data submission. Briefly, to prepare your data, users need to provide information for "Metadata", "Transformation/Construct/gRNA", and "Plant mutants" using a a template Excel file.
5. Additional links
iTAK
iTAK is a website that provides extensive information about plant transcription factors (TFs), transcriptional regulators (TRs) and protein kinases (PKs) from protein or nucleotide sequences and then classify individual TFs, TRs and PKs into different gene families.
TFGD (Tomato Functional Genomics database)
TEA (Tomato Expression Atlas)
SGN (Sol Genomics Network)
Addgene
Addgene is a non-profit plasmid repository. Addgene facilitates the exchange of genetic material between laboratories by offering plasmids and their associated cloning data to not-for-profit laboratories around the world. Addgene provides a free online database of plasmid cloning information and references, including lists of commonly used vector backbones, popular lentiviral plasmids and molecular cloning protocols.
This database is supported by NSF (IOS-1546625) and hosted by BTI.

